The data structure is based on xarray.Dataset¶
PIV data requires: - data in 2D or 3D matrices - coordinates for x,y or x,y,z - metadata that will contain the information from the header, information about the origin of the data file (image, experimental settings), units for each variables, coordinates, etc.
Among various possibilities the most suitable one is xarray, or so-called N-D labeled arrays, Read more about this format in this paper or in their docs
[6]:
import xarray as xr
[7]:
x = np.linspace(32.0, 128.0, 3) # 3 columns
y = np.linspace(16.0, 128.0, 4) # 4 rows
xm, ym = np.meshgrid(x, y)
u = np.ones_like(xm.T) + np.linspace(0.0, 7.0, 4)
v = (
np.zeros_like(ym.T)
+ np.linspace(0.0, 1.0, 4)
+ np.random.rand(3, 1)
- 0.5
)
u = u[:, :, np.newaxis]
v = v[:, :, np.newaxis]
chc = np.ones_like(u)
# plt.quiver(xm.T,ym.T,u,v)
u = xr.DataArray(
u, dims=("x", "y", "t"), coords={"x": x, "y": y, "t": [0]}
)
v = xr.DataArray(
v, dims=("x", "y", "t"), coords={"x": x, "y": y, "t": [0]}
)
chc = xr.DataArray(
chc, dims=("x", "y", "t"), coords={"x": x, "y": y, "t": [0]}
)
data = xr.Dataset({"u": u, "v": v, "chc": chc})
data.attrs["variables"] = ["x", "y", "u", "v"]
data.attrs["units"] = ["pix", "pix", "pix/dt", "pix/dt"]
data.attrs["dt"] = 1.0
data.attrs["files"] = ""
data
[7]:
<xarray.Dataset>
Dimensions: (x: 3, y: 4, t: 1)
Coordinates:
* x (x) float64 32.0 80.0 128.0
* y (y) float64 16.0 53.33 90.67 128.0
* t (t) int64 0
Data variables:
u (x, y, t) float64 1.0 3.333 5.667 8.0 1.0 ... 1.0 3.333 5.667 8.0
v (x, y, t) float64 0.1297 0.463 0.7963 1.13 ... 0.2745 0.6078 0.9412
chc (x, y, t) float64 1.0 1.0 1.0 1.0 1.0 1.0 1.0 1.0 1.0 1.0 1.0 1.0
Attributes:
variables: ['x', 'y', 'u', 'v']
units: ['pix', 'pix', 'pix/dt', 'pix/dt']
dt: 1.0
files: [8]:
### Using xarray plotting machinery
[9]:
import numpy as np
import xarray as xr
ds = xr.Dataset()
ds.coords["x"] = ("x", np.arange(10))
ds.coords["y"] = ("y", np.arange(20))
ds.coords["t"] = ("t", np.arange(4))
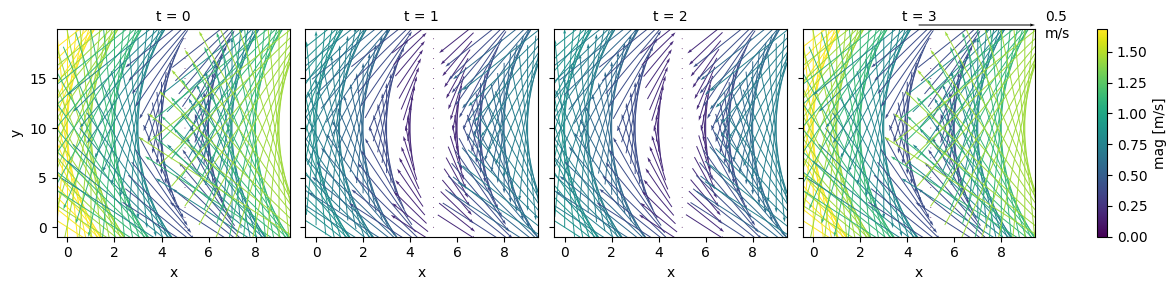
ds["u"] = np.sin((ds.x - 5) / 5) * np.sin((ds.y - 10) / 10)
ds["v"] = np.sin((ds.x - 5) / 5) * np.cos((ds.y - 10) / 10)
ds = ds * 2*np.cos((ds.t) * 2 * 3.14 /0.75)
ds["u"].attrs["units"] = "m/s"
ds["mag"] = np.hypot(ds.u, ds.v)
ds.mag.plot(col="t", x="x")
fg = ds.plot.quiver(x="x", y="y", u="u", v="v", col="t", hue="mag", scale=1)
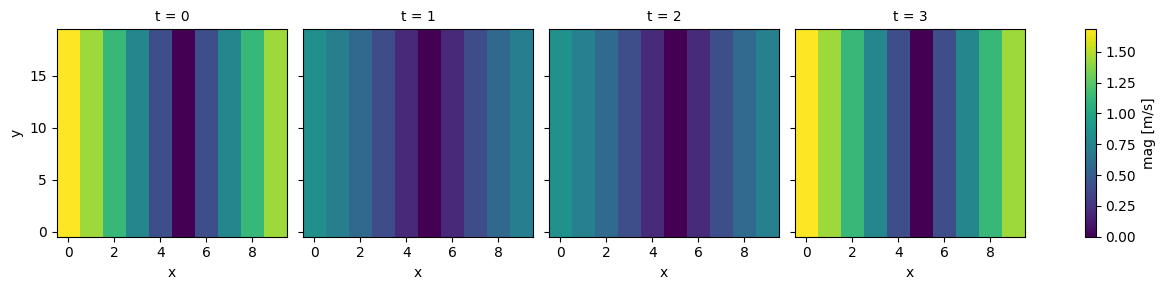
[16]:
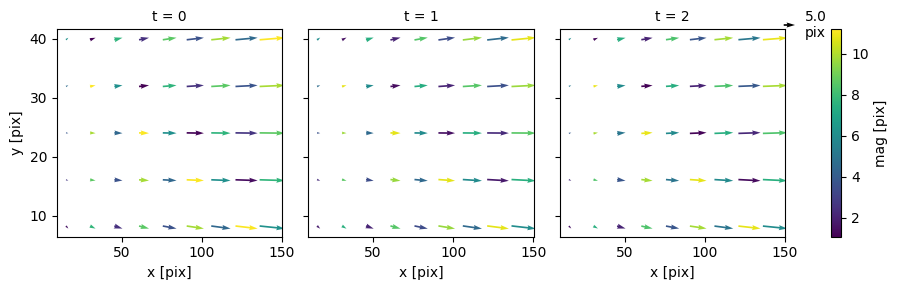
from pivpy import io, pivpy, graphics
ds = io.create_sample_Dataset(n_frames=3,rows=5,cols=9,noise_sigma=0.2)
ds["mag"] = np.hypot(ds.u, ds.v)
ds.plot.quiver(x="x", y="y", u="u", v="v", col="t", hue="mag", scale=100)
[16]:
<xarray.plot.facetgrid.FacetGrid at 0x7f755c399cd0>